All papers that have not been peer-reviewed will not appear here, including preprints. You can access my all of papers at 🔗Google Scholar.
2025
SC-AGR:Spatially-Constrained Attention for Context-Aware Graph Representation in Histopathology Whole Slide Image Analysis
Chengfei Cai, Jiahao Chen, Mingxin Liu, Jun Li, Jun Xu†(† corresponding author)
International Conference on Intelligent Computing (ICIC) 2025 Conference
Whole-slide images (WSIs) are widely used in histopathological examination, but current multi-instance learning (MIL) methods often fail to capture the complex interactions between instances in a WSI. Existing graph-based approaches, while modeling spatial relationships, are limited in their ability to capture interactions across arbitrary distances. We proposed a Spatially-Constrained Attention for Context-Aware Graph Representation framework (SC-AGR) to address these limitations. Specifically: 1) We introduce a context-aware graph representation that dynamically constructs spatial associations between different regions in a WSI, thereby more accurately capturing the characteristics of lesion tissue; 2) A spatially constrained attention mechanism enhances feature learning by aggregating adjacent nodes, allowing key patches to propagate information and improve WSI analysis; 3) We combined graph convolutional network (GCN) layers with instance clustering to further refine and constrain the graph representation feature space, boosting the model’s performance and data efficiency. Extensive experiments conducted on three public datasets demonstrate that SC-AGR outperformed state-of-the-art (SOTA) WSI analysis methods.


Classification of Whole-Slide Pathology Images Based on State Space Models and Graph Neural Networks
Feng Ding, Chengfei Cai, Jun Li, Mingxin Liu, Yiping Jiao, Zhengcan Wu, Jun Xu†(† corresponding author)
Electronics 2025 Journal
This work proposes a novel MIL framework: Dynamic Graph and State Space Model-Based MIL (DG-SSM-MIL). DG-SSM-MIL combines graph neural networks and selective state space models, leveraging the former’s ability to extract local and spatial features and the latter’s advantage in comprehensively understanding long-sequence instances. This enhances the model’s performance in diverse instance classification, improves its capability to handle long-sequence data, and increases the precision and scalability of feature fusion. We propose the Dynamic Graph and State Space Model (DynGraph-SSM) module, which aggregates local and spatial information of image patches through directed graphs and learns global feature representations using the Mamba model.


IBDAIM:Artificial intelligence for analyzing intestinal biopsies pathological images for assisted integrated diagnostic of inflammatory bowel disease
Chengfei Cai, Qianyun Shi, Mingxin Liu, Jun Li, Yangshu Zhou, Andi Xu, Dan Zhang, Yiping Jiao, Yao Liu, Xiaobin Cui, Jun Chen, Jun Xu†, Qi Sun†(† corresponding author)
International Journal of Medical Informatics 2025 Journal
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is challenging to diagnose accurately from pathological images due to its complex histological features. This study aims to develop an artificial intelligence (AI) model, IBDAIM, to assist pathologists in quickly and accurately diagnosing IBD by analyzing whole-slide images (WSIs) of intestinal biopsies.

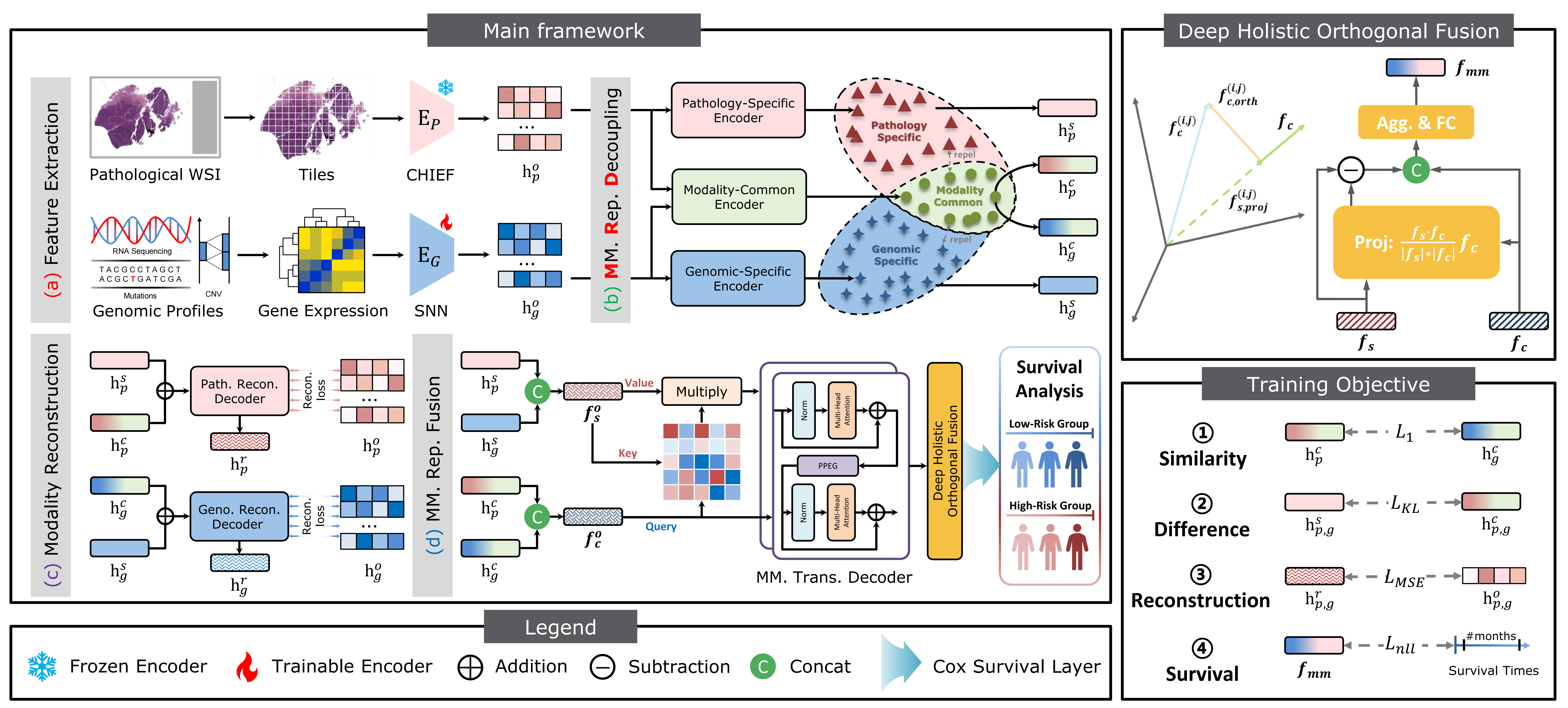
MurreNet:Modeling Holistic Interactions Between Histopathology and Genomic Profiles for Survival Prediction
Mingxin Liu, Chengfei Cai, Jun Li, Pengbo Xu, Jinze Li, Jiquan Ma, Jun Xu†(† corresponding author)
28th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2025 Conference
This paper presents a Multimodal Representation Decoupling Network (MurreNet) to advance cancer survival analysis. Specifically, we first propose a Multimodal Representation Decomposition (MRD) module to explicitly decompose paired input data into modality-specific and modality-shared representations, thereby reducing redundancy between modalities. Furthermore, the disentangled representations are further refined then updated through a novel training regularization strategy that imposes constraints on distributional similarity, difference, and representativeness of modality features. Finally, the augmented multimodal features are integrated into a joint representation via proposed Deep Holistic Orthogonal Fusion (DHOF) strategy. Extensive experiments conducted on six TCGA cancer cohorts demonstrate that our MurreNet achieves state-of-the-art (SOTA) performance in survival prediction.

2024
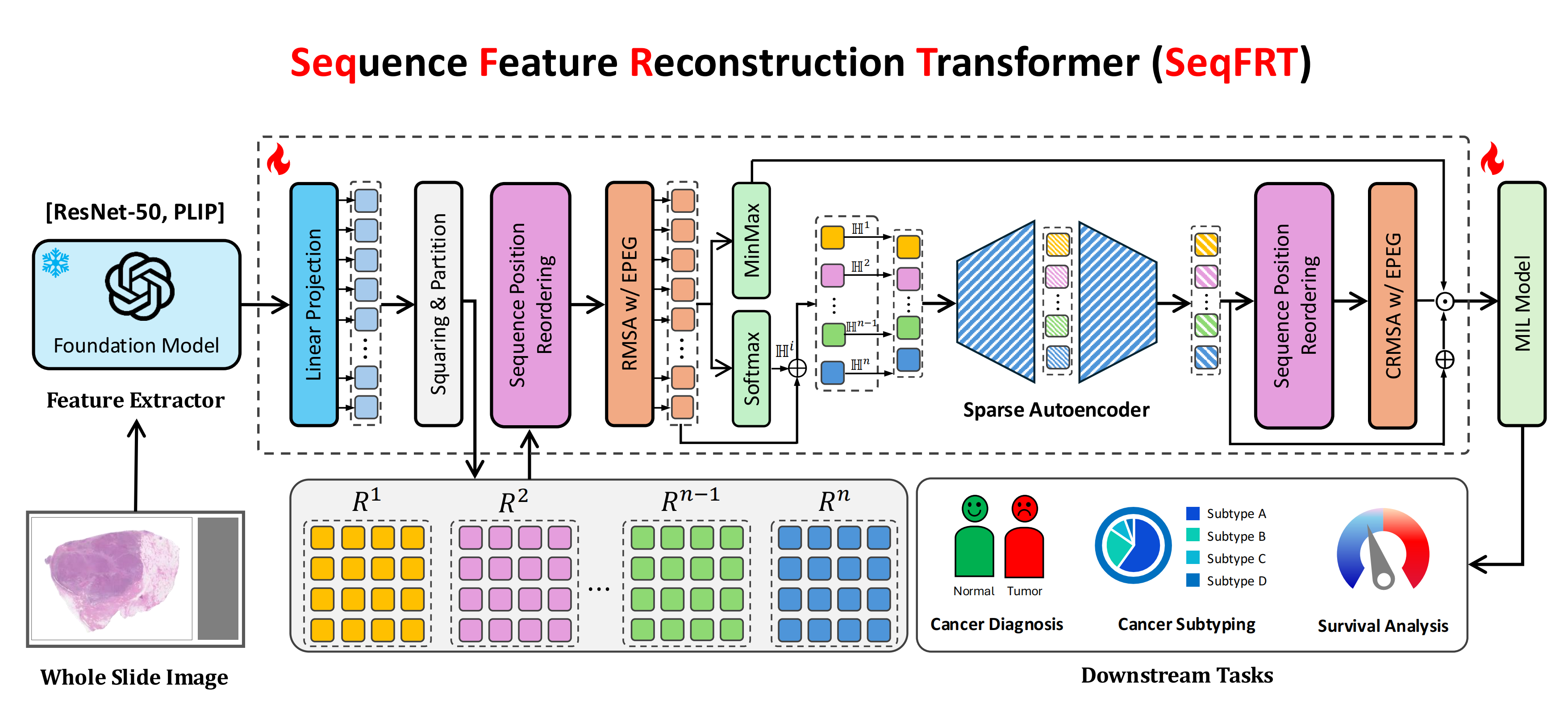
SeqFRT: Towards Effective Adaption of Foundation Model via Sequence Feature Reconstruction in Computational Pathology
Chengfei Cai, Jun Li, Mingxin Liu, Yiping Jiao, Jun Xu†(† corresponding author)
2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2024 ConferenceOral
In this paper, we present an innovative weakly-supervised sequence feature optimization method to solve the problem of sub-optimal feature extraction by the foundation model in the traditional MIL paradigm. The proposed SeqFRT leverages a sequence position optimization strategy to exploit the inherent valuable information embedded within the long pathological feature sequences and a sequence sparsity enhancement technique to highly enhance the ability to discriminate and extract the latent representations instead of redundant information, leading to preserving essential information for reconstructing input pathological sequence representation which is crucial for downstream tasks in computational pathology.

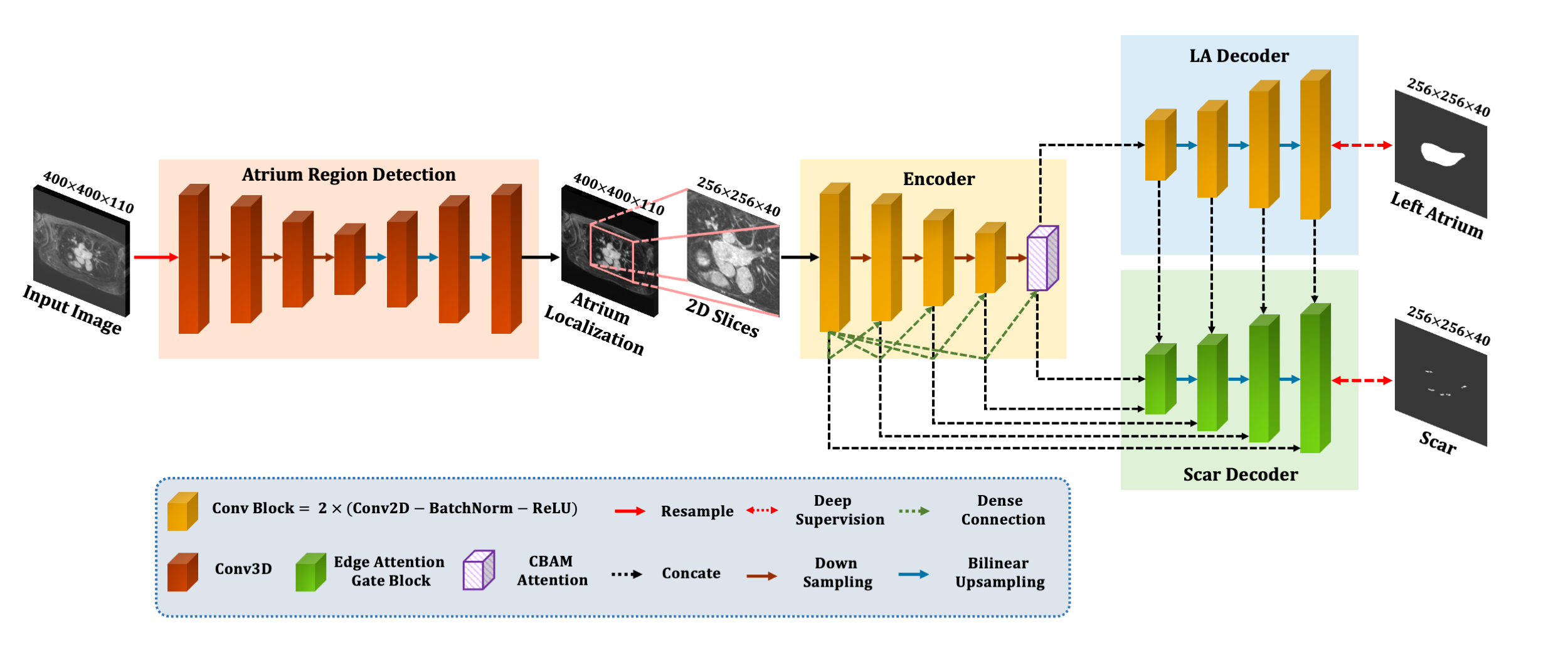
Edge and dense attention U-net for atrial scar segmentation in LGE-MRI
Gaoyuan Li, Mingxin Liu, Jun Lu, Jiquan Ma†(† corresponding author)
Biomedical Physics & Engineering Express 2024 Journal
We introduce a dual branch network, incorporating edge attention, and deep supervision strategy. Edge attention is introduced to fully utilize the spatial relationship between the scar and the atrium. Besides, dense attention is embedded in the bottom layer to solve feature disappearance. At the same time, deep supervision accelerates the convergence of the model and improves segmentation accuracy.

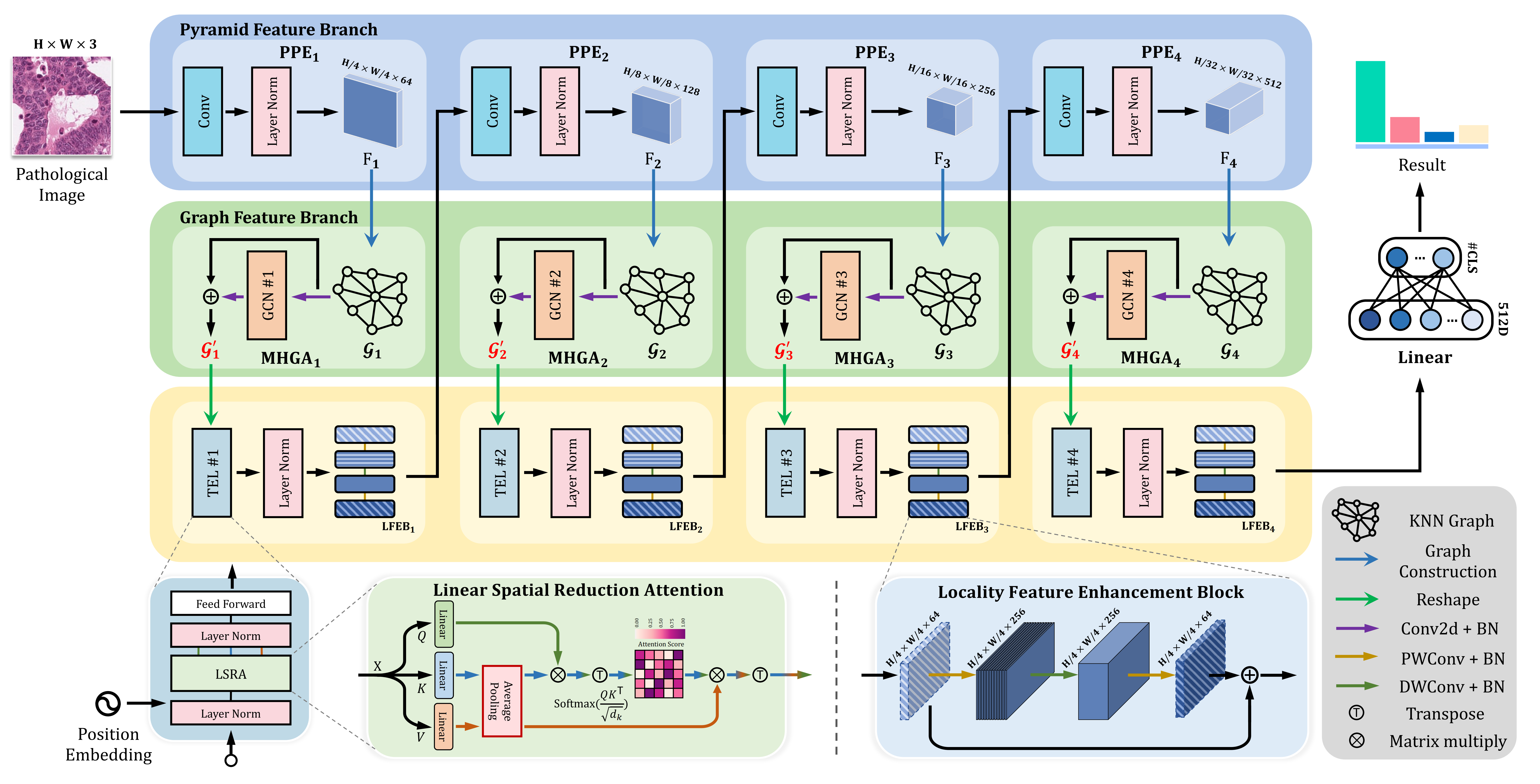
Exploiting Geometric Features via Hierarchical Graph Pyramid Transformer for Cancer Diagnosis Using Histopathological Images
Mingxin Liu, Yunzan Liu, Pengbo Xu, Hui Cui, Jing Ke, Jiquan Ma†(† corresponding author)
IEEE Transactions on Medical Imaging 2024 Journal
This study proposed HGPT, a novel framework that jointly considers geometric and global representation for cancer diagnosis in histopathological images. HGPT leverages a multi-head graph aggregator to aggregate the geometric representation from pathological morphological features, and a locality feature enhancement block to highly enhance the 2D local feature perception in vision transformers, leading to improved performance on histopathological image classification. Extensive experiments on Kather-5K, MHIST, NCT-CRC-HE, and GasHisSDB four public datasets demonstrate the advantages of the proposed HGPT over bleeding-edge approaches in improving cancer diagnosis performance.

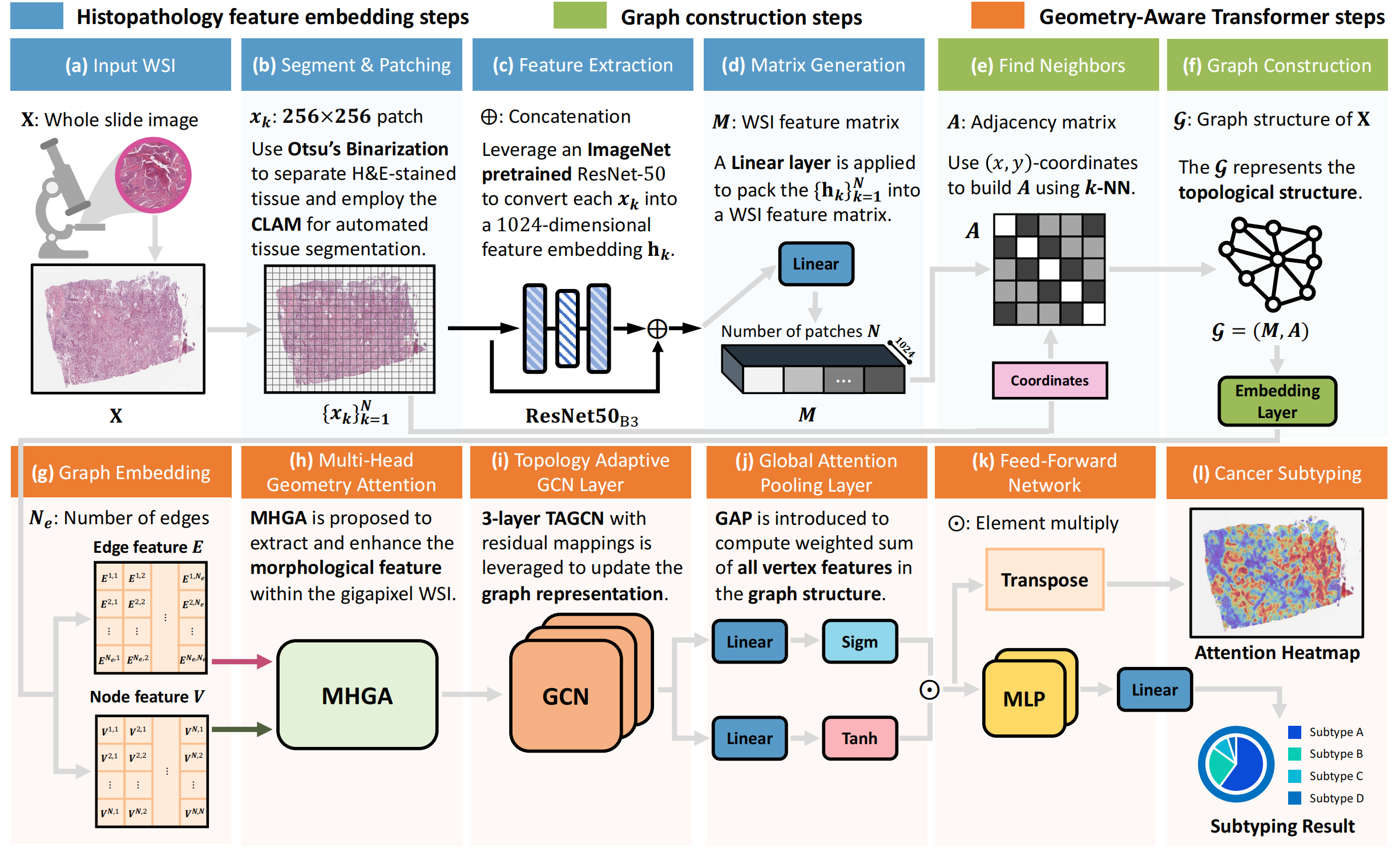
Unleashing the Infinity Power of Geometry:A Novel Geometry-Aware Transformer (GOAT) for Whole Slide Histopathology Image Analysis
Mingxin Liu, Yunzan Liu, Pengbo Xu, Jiquan Ma†(† corresponding author)
2024 IEEE International Symposium on Biomedical Imaging (ISBI) 2024 ConferenceOral
We proposed a novel weakly-supervised framework, Geometry-Aware Transformer (GOAT), in which we urge the model to pay attention to the geometric characteristics within the tumor microenvironment which often serve as potent indicators. In addition, a context-aware attention mechanism is designed to extract and enhance the morphological features within WSIs. Extensive experimental results demonstrated that the proposed method is capable of consistently reaching superior classification outcomes for gigapixel whole slide images.

2023
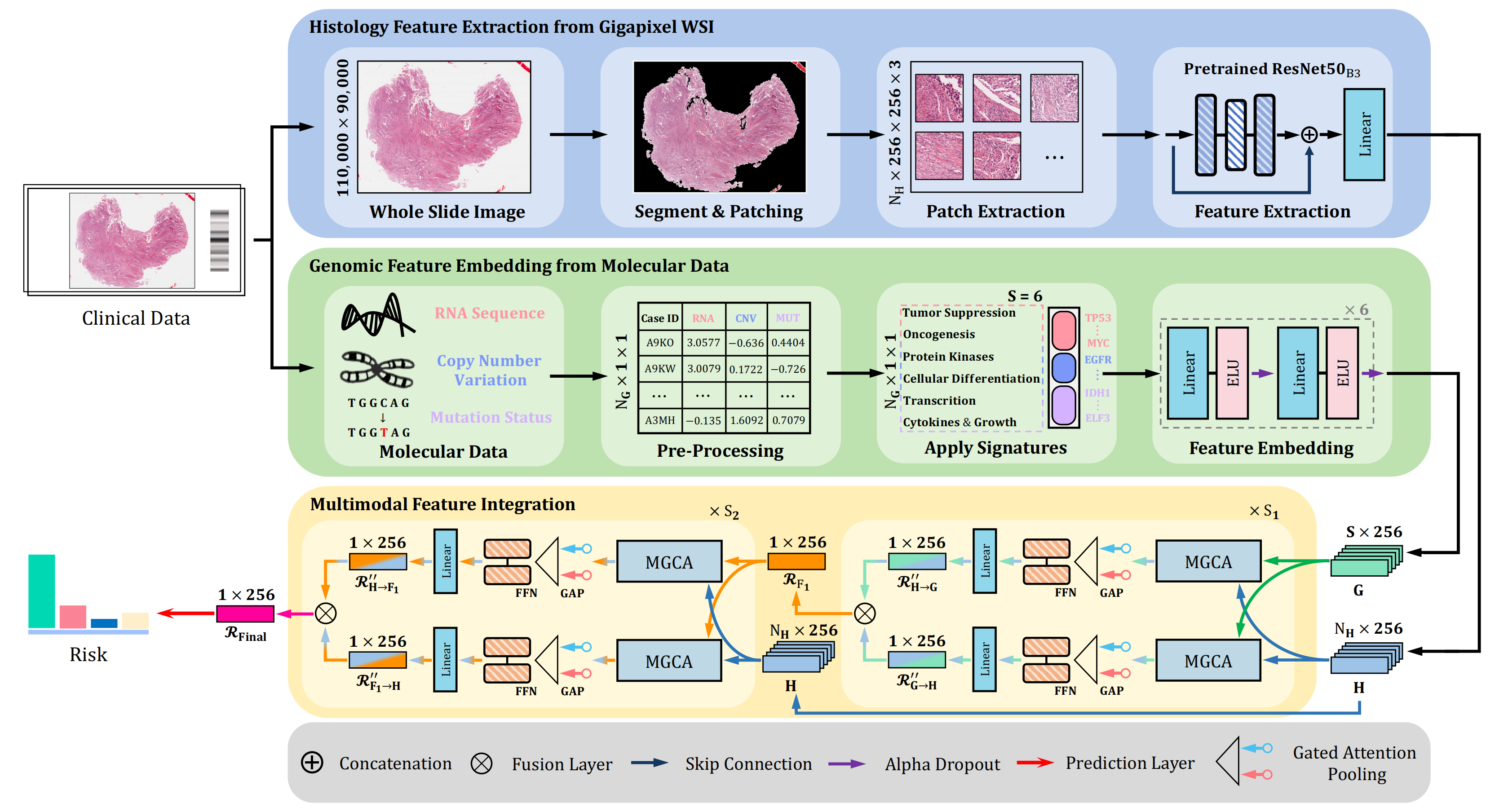
MGCT:Mutual-Guided Cross-Modality Transformer for Survival Outcome Prediction using Integrative Histopathology-Genomic Features
Mingxin Liu, Yunzan Liu, Hui Cui, Chunquan Li†, Jiquan Ma†(† corresponding author)
2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2023 ConferenceOral
We propose the Mutual-Guided Cross-Modality Transformer (MGCT), a weakly-supervised, attention-based multimodal learning framework that can combine histology features and genomic features to model the genotype-phenotype interactions within the tumor microenvironment. Extensive experimental results on five benchmark datasets consistently emphasize that MGCT outperforms the state-of-the-art (SOTA) methods.
