Mingxin Liu
 Ph.D. student, Nanjing University of Information Science and Technology
Ph.D. student, Nanjing University of Information Science and Technology
I am a first year Ph.D. student advised by Prof. Jun Xu at Jiangsu Key Laboratory of Intelligent Medical Image Computing (IMIC), Nanjing University of Information Science and Technology. My research focuses on Computational Pathology, utilizing weakly-supervised learning, geometric deep learning, and multimodal learning techniques. Prior to starting my Ph.D. study, I obtained my M.S degree in Computer Science at Heilongjiang University, advised by Prof. Jiquan Ma, and worked with Prof. Dinggang Shen, Prof. Jing Ke, Prof. Chunquan Li, and Dr. Hui Cui.

Education
-

Nanjing University of Information Science and Technology
Ph.D. student in Artificial Intelligence Sep. 2024 - Present
-

Heilongjiang University
M.S. in Computer Science Sep. 2021 - Jul. 2024
-

Heilongjiang International University
B.S. in Computer Science and Technology Sep. 2017 - Jul. 2021
Service
- Reviewer of Engineering Applications of Artificial Intelligence (EAAI, since 2025)
- Reviewer of IEEE Transactions on Medical Imaging (TMI, since 2025)
- Reviewer of International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2025/2026)
- Reviewer of International Joint Conference on Neural Networks (IJCNN 2025)
- Reviewer of Digital Signal Processing (DSP, since 2025)
- Reviewer of Computerized Medical Imaging and Graphics (CMIG, since 2024)
- Reviewer of Neural Information Processing Systems (NeurIPS) 2022 Cell Segmentation Challenge
Honors & Awards
- Outstanding Master’s Degree Thesis in Heilongjiang University 2024
- Second-Class Graduate Scholarship in Heilongjiang University 2023
- First-Class Graduate Scholarship in Heilongjiang University 2022
- First-Class Graduate Scholarship in Heilongjiang University 2021
Selected Publications (view all )

Data- and Knowledge-Driven Multimodal Learning in Computational Pathology:A Comprehensive Survey
Mingxin Liu, Chengfei Cai, Depin Chen, Jun Li, Jinze Li, Wenlong Ming, Jun Xu†(† corresponding author)
EngMedicine 2026 Journal
Computational pathology (CPath) is undergoing a transformative evolution, driven by the convergence of large-scale biomedical data and breakthroughs in deep learning and large language models. Medical data is collected using different measurement modalities or sources, each providing complementary information. Multi-scale and multi-modal medical data represent information across multiple biological and clinical levels, ranging from molecular measurements to whole-organ and patient-level observations. While most efforts in the field of CPath focused on unimodal analysis of whole slide images (WSIs), the field has expanded toward multimodal integration, combining pathological WSIs with radiology imaging, genomic profiles, and Electronic Health Records to achieve comprehensive and context-aware interpretation of disease phenotypes. In this survey, we synthesize reviews of advances in CPath along two axes: data-driven multimodal learning and knowledge-driven enhancement. We also discuss the fusion strategies across visual and non-visual modalities, cross-modal contrastive learning, and self-supervised pretraining with unannotated data at scale. In parallel, we highlight representative applications that incorporate prior domain knowledge, such as morphological constraints, geometrical representations, and text prompts, into model design and supervision to enhance both generalizability and interpretability. Finally, we examine transformative clinical applications and highlight future directions toward generalizable, explainable, and clinically actionable artificial intelligence for pathology systems. By bridging cutting-edge technical innovation with real-world diagnostic requirements, in conducting this review, we aim to provide a conceptual roadmap for the next generation of trustworthy artificial intelligence in digital pathology.

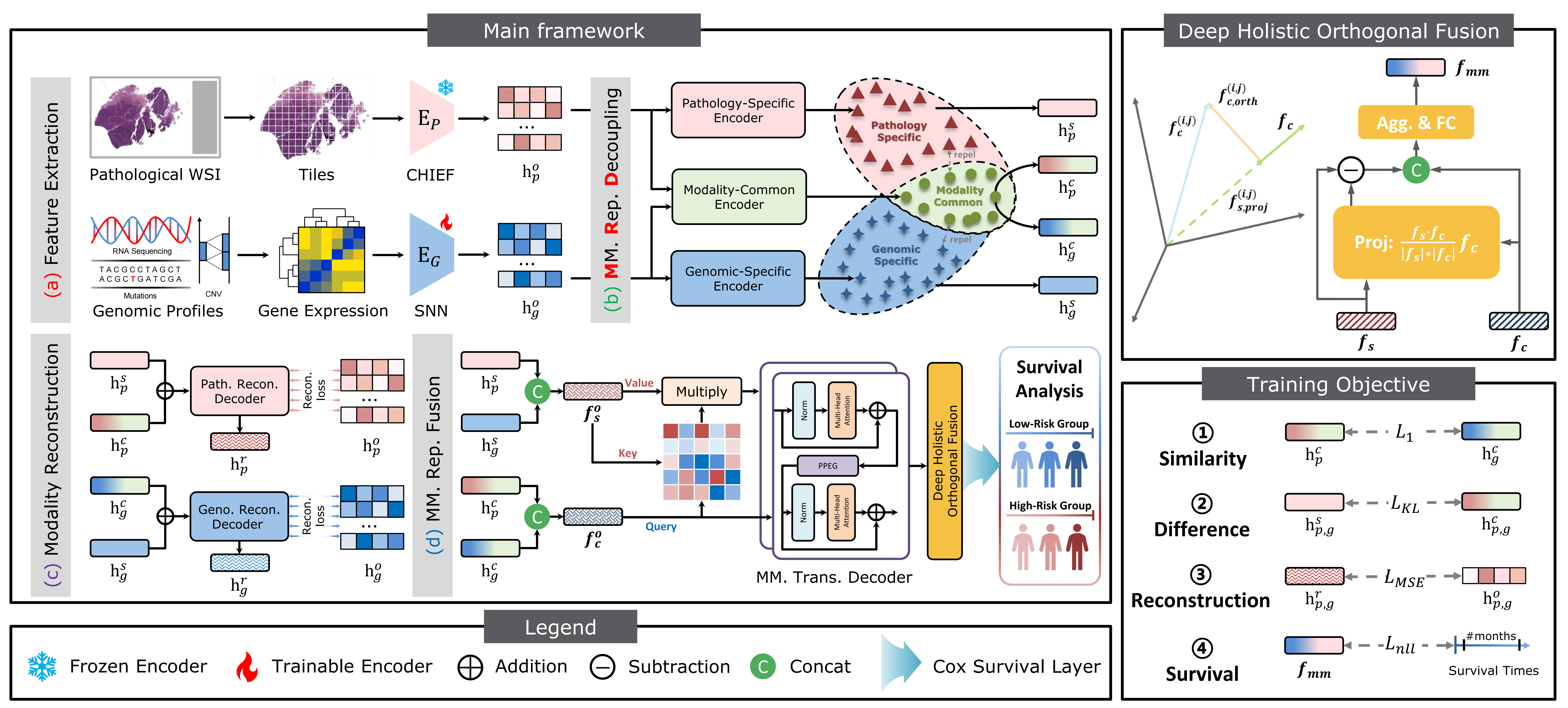
MurreNet:Modeling Holistic Interactions Between Histopathology and Genomic Profiles for Survival Prediction
Mingxin Liu, Chengfei Cai, Jun Li, Pengbo Xu, Jinze Li, Jiquan Ma, Jun Xu†(† corresponding author)
28th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2025 Conference
This paper presents a Multimodal Representation Decoupling Network (MurreNet) to advance cancer survival analysis. Specifically, we first propose a Multimodal Representation Decomposition (MRD) module to explicitly decompose paired input data into modality-specific and modality-shared representations, thereby reducing redundancy between modalities. Furthermore, the disentangled representations are further refined then updated through a novel training regularization strategy that imposes constraints on distributional similarity, difference, and representativeness of modality features. Finally, the augmented multimodal features are integrated into a joint representation via proposed Deep Holistic Orthogonal Fusion (DHOF) strategy. Extensive experiments conducted on six TCGA cancer cohorts demonstrate that our MurreNet achieves state-of-the-art (SOTA) performance in survival prediction.

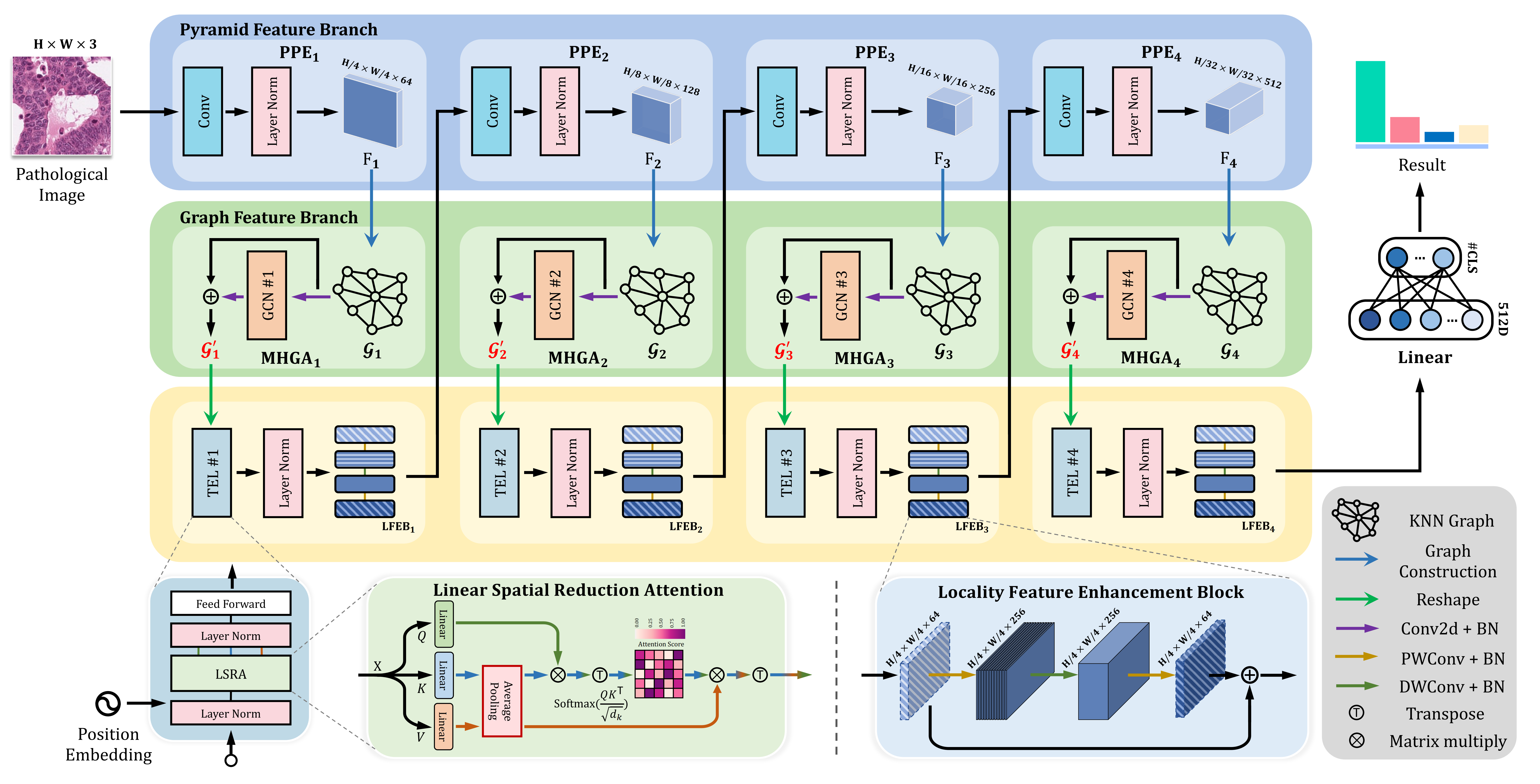
Exploiting Geometric Features via Hierarchical Graph Pyramid Transformer for Cancer Diagnosis Using Histopathological Images
Mingxin Liu, Yunzan Liu, Pengbo Xu, Hui Cui, Jing Ke, Jiquan Ma†(† corresponding author)
IEEE Transactions on Medical Imaging 2024 Journal
This study proposed HGPT, a novel framework that jointly considers geometric and global representation for cancer diagnosis in histopathological images. HGPT leverages a multi-head graph aggregator to aggregate the geometric representation from pathological morphological features, and a locality feature enhancement block to highly enhance the 2D local feature perception in vision transformers, leading to improved performance on histopathological image classification. Extensive experiments on Kather-5K, MHIST, NCT-CRC-HE, and GasHisSDB four public datasets demonstrate the advantages of the proposed HGPT over bleeding-edge approaches in improving cancer diagnosis performance.

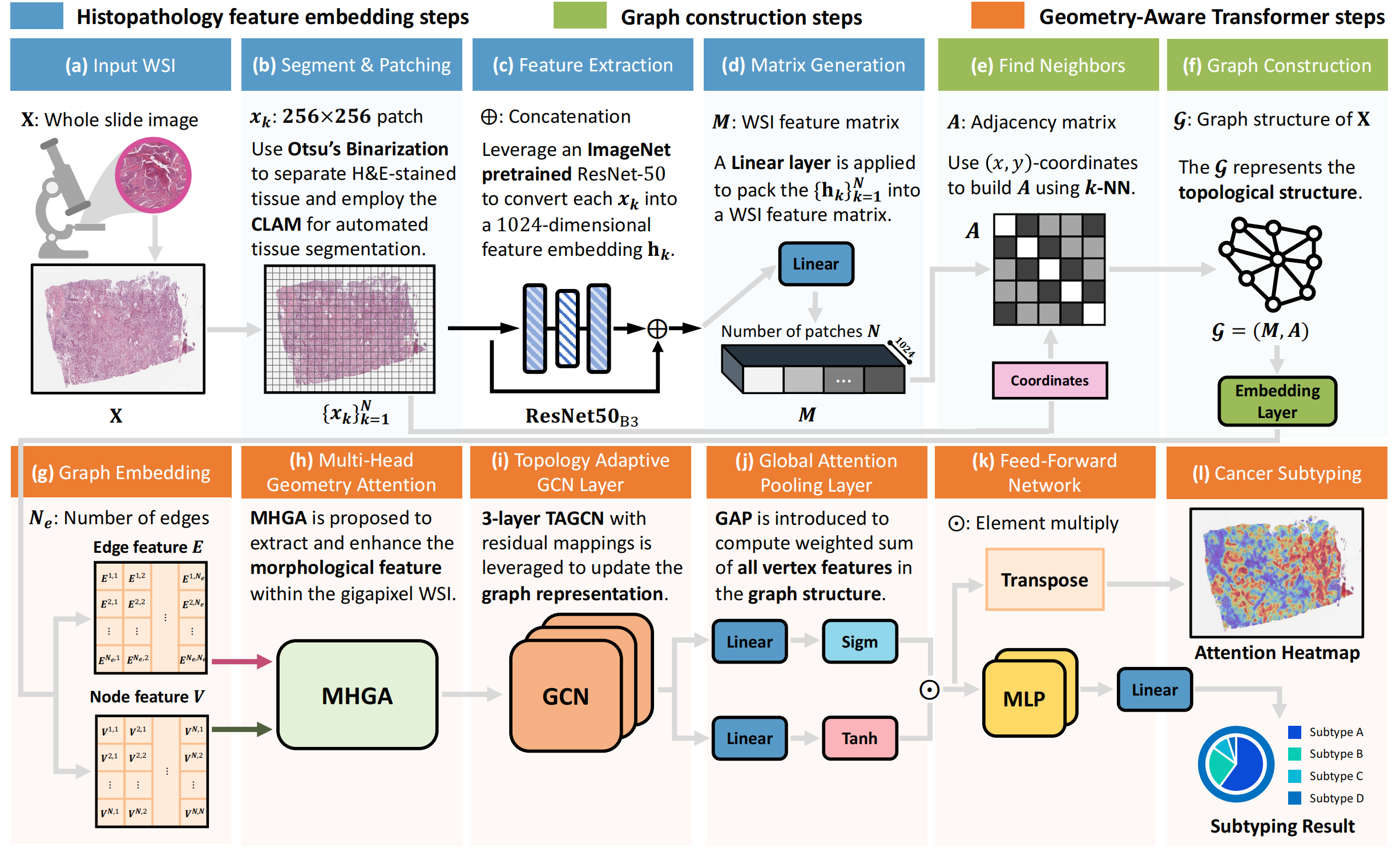
Unleashing the Infinity Power of Geometry:A Novel Geometry-Aware Transformer (GOAT) for Whole Slide Histopathology Image Analysis
Mingxin Liu, Yunzan Liu, Pengbo Xu, Jiquan Ma†(† corresponding author)
2024 IEEE International Symposium on Biomedical Imaging (ISBI) 2024 ConferenceOral
We proposed a novel weakly-supervised framework, Geometry-Aware Transformer (GOAT), in which we urge the model to pay attention to the geometric characteristics within the tumor microenvironment which often serve as potent indicators. In addition, a context-aware attention mechanism is designed to extract and enhance the morphological features within WSIs. Extensive experimental results demonstrated that the proposed method is capable of consistently reaching superior classification outcomes for gigapixel whole slide images.

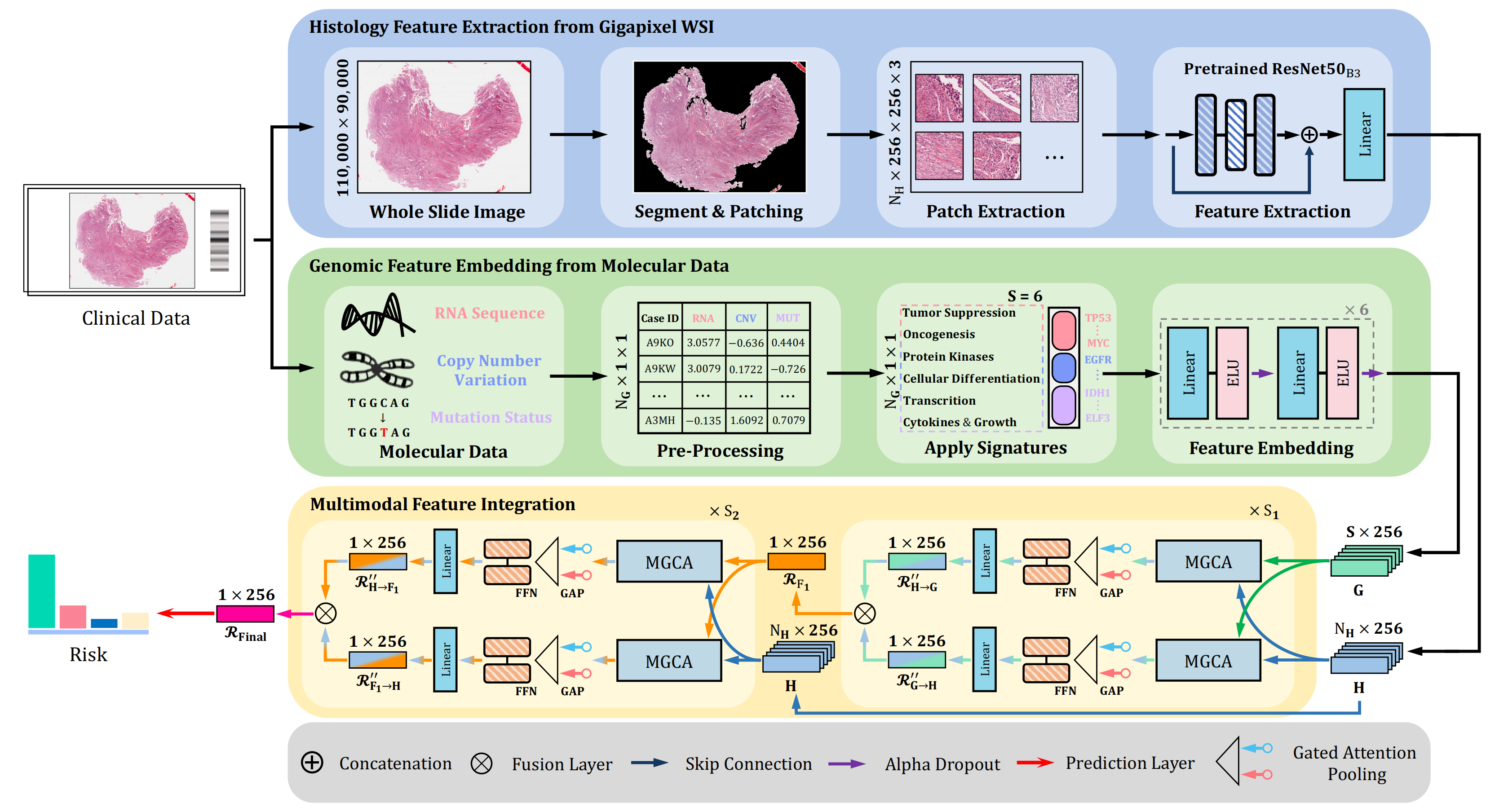
MGCT:Mutual-Guided Cross-Modality Transformer for Survival Outcome Prediction using Integrative Histopathology-Genomic Features
Mingxin Liu, Yunzan Liu, Hui Cui, Chunquan Li†, Jiquan Ma†(† corresponding author)
2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2023 ConferenceOral
We propose the Mutual-Guided Cross-Modality Transformer (MGCT), a weakly-supervised, attention-based multimodal learning framework that can combine histology features and genomic features to model the genotype-phenotype interactions within the tumor microenvironment. Extensive experimental results on five benchmark datasets consistently emphasize that MGCT outperforms the state-of-the-art (SOTA) methods.
